NOTE: This vignette uses outdated versions of stan syntax and the chkptstanr package. It will be updated shortly.
The following examples walk through using chkptstanr with the Stan
The basic idea is to (1) write a custom Stan model
(done by the user), (2) fit the model with cmdstanr (with
the desired number of checkpoints), and then (3) return a
cmststanr object. All but step (1) is done internally, so
the workflow is very similar to using cmdstanr.
Example 1: Eight Schools
Storage
The initial overhead is to create a folder that will store the checkpoints, i.e.,
path <- create_folder(folder_name = "chkpt_folder_m1")Stan Model
Next is the Stan model:
stan_code <- "
data {
int<lower=0> n;
real y[n];
real<lower=0> sigma[n];
}
parameters {
real mu;
real<lower=0> tau;
vector[n] eta;
}
transformed parameters {
vector[n] theta;
theta = mu + tau * eta;
}
model {
target += normal_lpdf(eta | 0, 1);
target += normal_lpdf(y | theta, sigma);
}
"Stan Data
When using chkpt_stan(), this requires supplying a list
to the data argument, much like using rstan.
Model Fitting
2 Checkpoints
To show the basic idea of checkpointing, the following was stopped after 2 checkpoints.
fit_m1 <- chkpt_stan(model_code = stan_code,
data = stan_data,
iter_warmup = 1000,
iter_sampling = 1000,
iter_per_chkpt = 250,
path = path)
#> Compiling Stan program...
#> Initial Warmup (Typical Set)
#> Chkpt: 1 / 8; Iteration: 250 / 2000 (warmup)
#> Chkpt: 2 / 8; Iteration: 500 / 2000 (warmup)Finish Sampling
To finish the remaining 6 checkpoints run the same code, i.e.,
fit_m1 <- chkpt_stan(model_code = stan_code,
data = stan_data,
iter_warmup = 1000,
iter_sampling = 1000,
iter_per_chkpt = 250,
path = path)
#> Sampling next checkpoint
#> Chkpt: 3 / 8; Iteration: 750 / 2000 (warmup)
#> Chkpt: 4 / 8; Iteration: 1000 / 2000 (warmup)
#> Chkpt: 5 / 8; Iteration: 1250 / 2000 (sample)
#> Chkpt: 6 / 8; Iteration: 1500 / 2000 (sample)
#> Chkpt: 7 / 8; Iteration: 1750 / 2000 (sample)
#> Chkpt: 8 / 8; Iteration: 2000 / 2000 (sample)
#> Checkpointing completeCombine Draws
Each checkpoint contains 250 draws from the posterior. These need to
be combined with combine_chkpt_draws(), i.e.,
draws <- combine_chkpt_draws(fit_m1)We developed chkptstanr to work seamlessly with the
Stan ecosystem. The object draws has been
constructed to mimic what is provided when using
cmdstanr directly.
combine_chkpt_draws(fit_m1)
#> # A draws_array: 1000 iterations, 2 chains, and 19 variables
#> , , variable = lp__
#>
#> chain
#> iteration 1 2
#> 1 -34 -43
#> 2 -37 -41
#> 3 -36 -39
#> 4 -38 -38
#> 5 -38 -41
#>
#> , , variable = mu
#>
#> chain
#> iteration 1 2
#> 1 5.2 2.6
#> 2 11.3 6.7
#> 3 -2.7 5.3
#> 4 -2.9 3.7
#> 5 -2.7 14.2
#>
#> , , variable = tau
#>
#> chain
#> iteration 1 2
#> 1 23.3 2.61
#> 2 6.7 0.21
#> 3 12.7 4.44
#> 4 21.1 7.29
#> 5 18.8 10.94
#>
#> , , variable = eta[1]
#>
#> chain
#> iteration 1 2
#> 1 0.10 -0.61
#> 2 0.89 -0.87
#> 3 1.62 0.83
#> 4 1.99 0.84
#> 5 -0.16 1.22
#>
#> # ... with 995 more iterations, and 15 more variablesSummary
draws can then be used with the R package
posterior
posterior::summarise_draws(draws)
#> # A tibble: 19 x 10
#> variable mean median sd mad q5 q95 rhat ess_bulk ess_tail
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 lp__ -39.5 -39.2 2.59 2.58 -44.2 -35.9 1.00 640. 1008.
#> 2 mu 7.77 7.92 5.48 5.10 -1.43 16.0 1.01 530. 325.
#> 3 tau 6.82 5.32 5.75 4.71 0.434 18.7 1.00 649. 658.
#> 4 eta[1] 0.383 0.413 0.929 0.909 -1.20 1.87 1.00 1650. 1233.
#> 5 eta[2] -0.00335 -0.00816 0.841 0.814 -1.34 1.40 1.00 1443. 1307.
#> 6 eta[3] -0.176 -0.174 0.931 0.906 -1.67 1.42 1.00 1829. 1424.
#> 7 eta[4] -0.00521 0.000856 0.862 0.841 -1.47 1.39 1.00 1565. 1407.
#> 8 eta[5] -0.312 -0.350 0.873 0.835 -1.72 1.24 1.00 1661. 1616.
#> 9 eta[6] -0.193 -0.190 0.889 0.909 -1.59 1.28 1.00 1915. 1404.
#> 10 eta[7] 0.387 0.358 0.876 0.864 -1.09 1.81 1.00 1574. 1370.
#> 11 eta[8] 0.0805 0.0611 0.970 0.960 -1.51 1.66 1.00 1031. 1236.
#> 12 theta[1] 11.5 10.2 8.29 6.99 0.268 26.4 1.00 1042. 728.
#> 13 theta[2] 7.87 7.87 6.20 5.66 -2.27 17.8 1.00 1549. 1515.
#> 14 theta[3] 6.01 6.63 8.25 6.63 -8.69 18.1 1.00 1102. 1075.
#> 15 theta[4] 7.75 7.76 6.65 5.96 -3.06 18.9 1.00 1674. 1210.
#> 16 theta[5] 5.05 5.70 6.44 5.75 -7.06 14.4 1.00 1405. 1416.
#> 17 theta[6] 6.21 6.60 6.92 6.15 -5.98 16.9 1.00 1890. 1195.
#> 18 theta[7] 10.8 10.1 6.71 6.03 0.992 23.1 1.00 1497. 1767.
#> 19 theta[8] 8.35 8.41 7.72 6.66 -3.88 20.7 1.00 1081. 1075.Visualization with bayesplot
The popular R package bayesplot can
also be used.
bayesplot::mcmc_trace(draws) +
geom_vline(xintercept = seq(0, 1000, 250),
alpha = 0.25,
size = 2)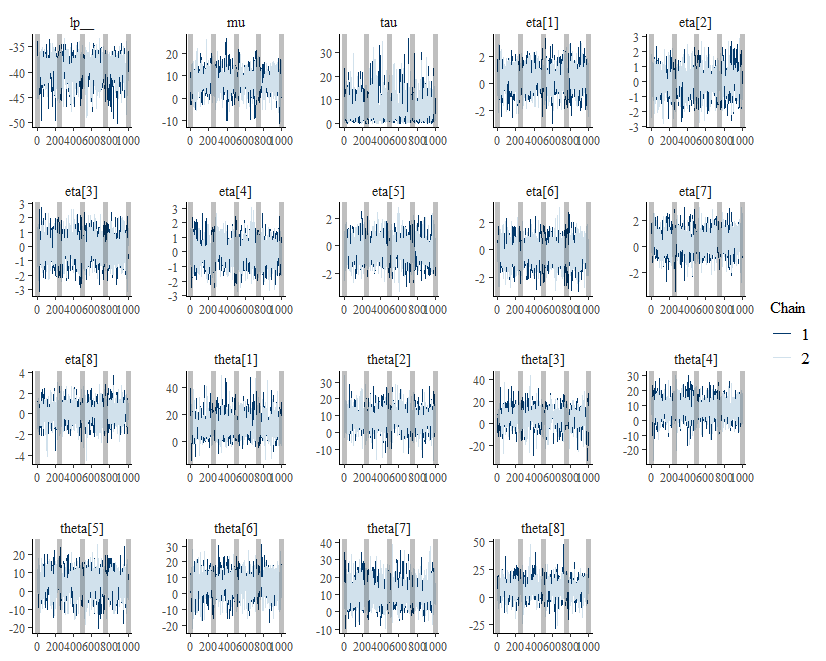
This vertical lines are placed at each checkpoint.